Applications in Nucleic Acid Therapeutics
Label-free Analysis of Nanomedicine
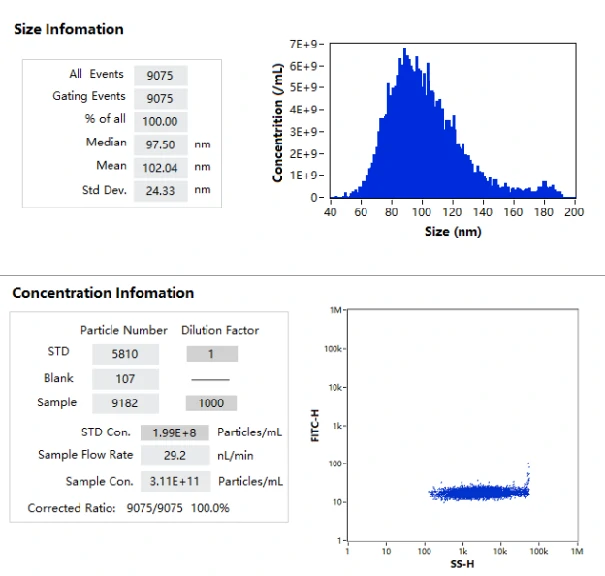
NanoFCM enables the label-free analysis of particle size distribution and particle concentration of lipid nanoparticles at single-particle level.
LNP Loading Fraction
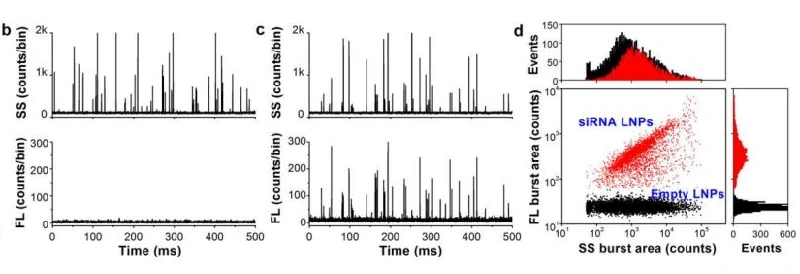
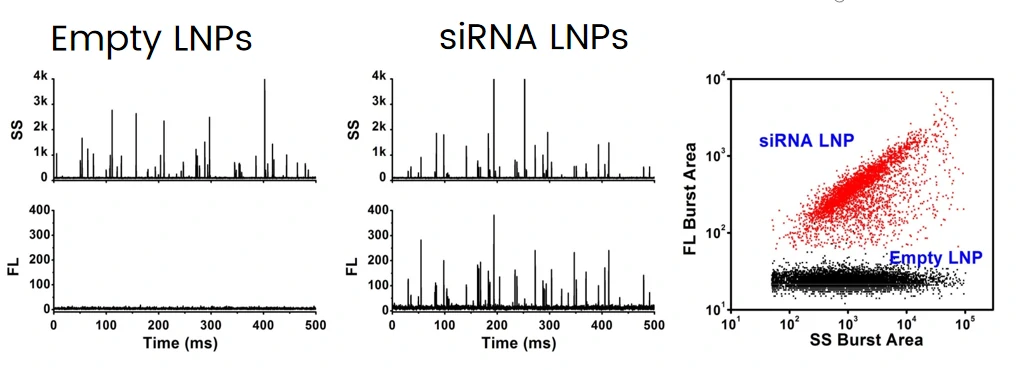
The excellent light scattering sensitivity of NanoFCM enables the identification of all lipid nanoparticles, based on the concurrent fluorescent detection, further evaluation of loading fraction and encapsulation efficiency can be achieved by nucleic acid staining. Lipid nanoparticles encapsulating siRNA are one of the most extensively clinically validated means of RNA interference. However, it is challenging to determine the precise particle count and the fraction of loaded particles. Hereon, NanoFCM was implemented to characterize siRNA-loaded LNPs and empty LNPs prepared by Alnylam Pharmaceuticals (Cambridge, MA, USA). Cell-permeant dye that labels nucleic acids was used to label the samples. As shown in Figure, near-complete discrimination between the empty and siRNA-loaded LNPs was achieved based on the fluorescence signals, and the fraction of siRNA loading was determined to be approximately 100%.
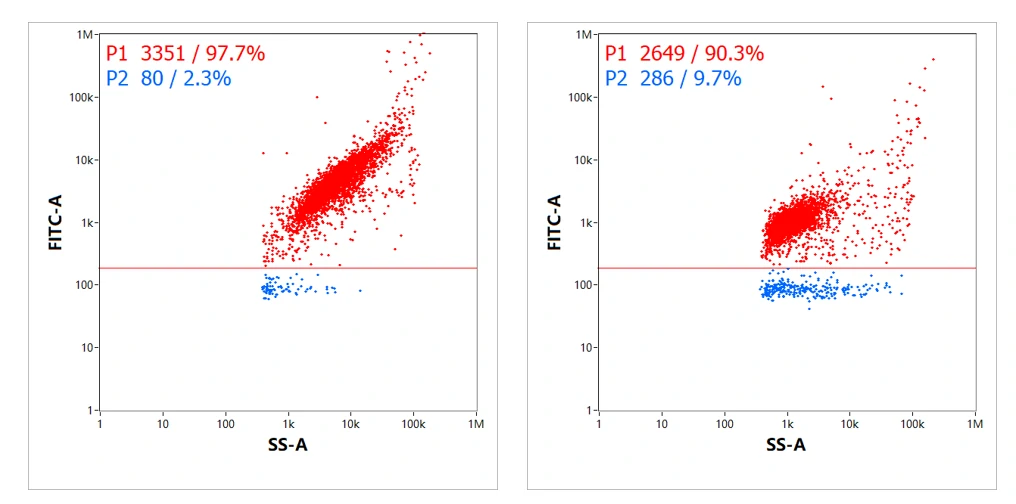
Besides, the loading fraction of two mRNA LNPs was evaluated using similar labeling strategy. The results indicated that 97.7% and 90.3% of LNPs were successfully loaded with mRNA, respectively. Given the real-time burst traces, dot-plot distribution and connections between fluorescence and side scatter provided by NanoFCM, detection of loading fraction solely is far from satisfying.
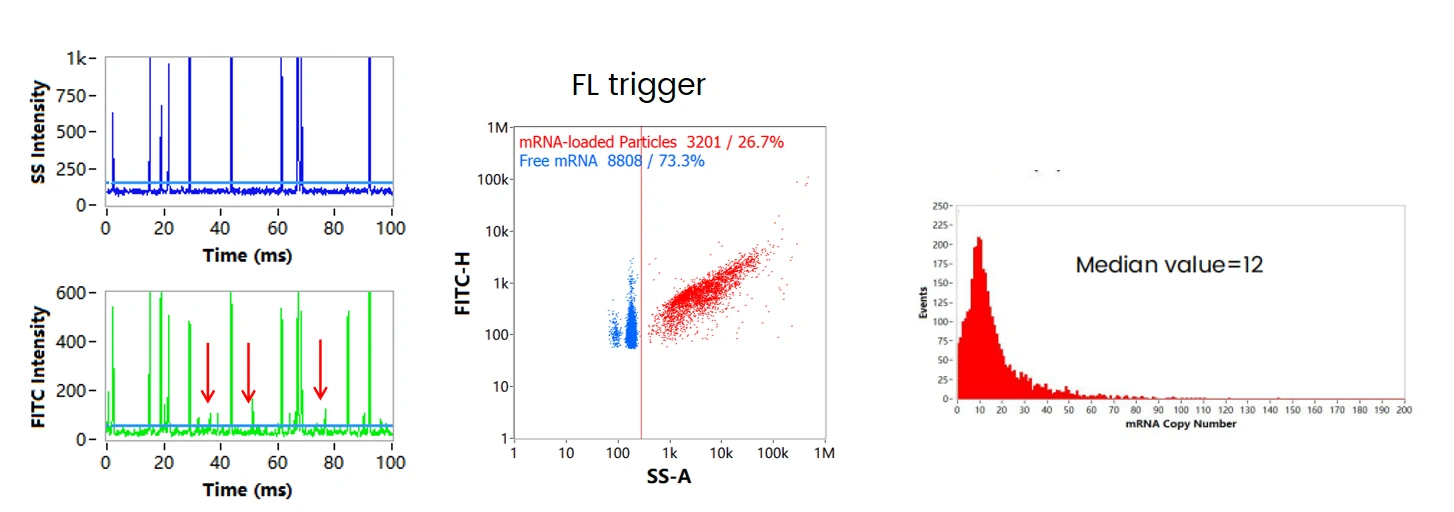
mRNA Copy Number and Encapsulation Efficiency
After labeling with cell-permeant nucleic acid stain, some fluorescent signals that are not associated with corresponding side scatter signals are observed, and the signal intensities (FL Intensity) are generally lower than the fluorescent signals that are associated with side scatter signals (shown in the arrows). These fluorescence signals are further confirmed to attribute to single free mRNAs by employing the pure mRNA molecules as control (data not shown). By choosing FL trigger, the free mRNA and mRNA encapusulated LNPs are shown in the dotplot. The mRNA copy number of each LNP can be achived by dividing the fluorescence intensities of each LNP by the intensity of free mRNA. The copy number ranges from 1 to around 100, with median value equals to 12. The encapsulation efficiency of mRNA can then be validated, which is 80.5% in this case.
Dual Trigger for mRNA Identification
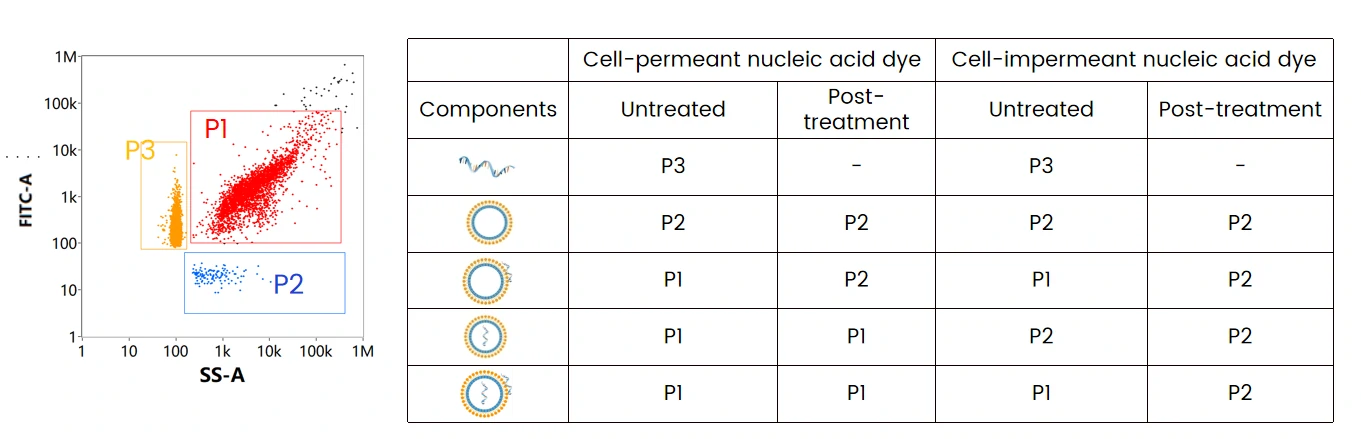
The mRNA LNP formulation mainly contains three components: mRNA encapsulated LNPs (P1), empty LNPs (P2), and free mRNA (P3). When labeled with cell-permeant nucleic acid dye, and through SS-FL dual trigger, the three components can be displayed and well resolved in the SS-FL dotplot. After treatment with nuclease, the free mRNA population disappeared, while the ratio of empty LNPs increased, which indicates the attachment of mRNA on the surface of some LNPs. This is further verified by labeling with cell-impermeant nucleic acid dye following nuclease treatment.
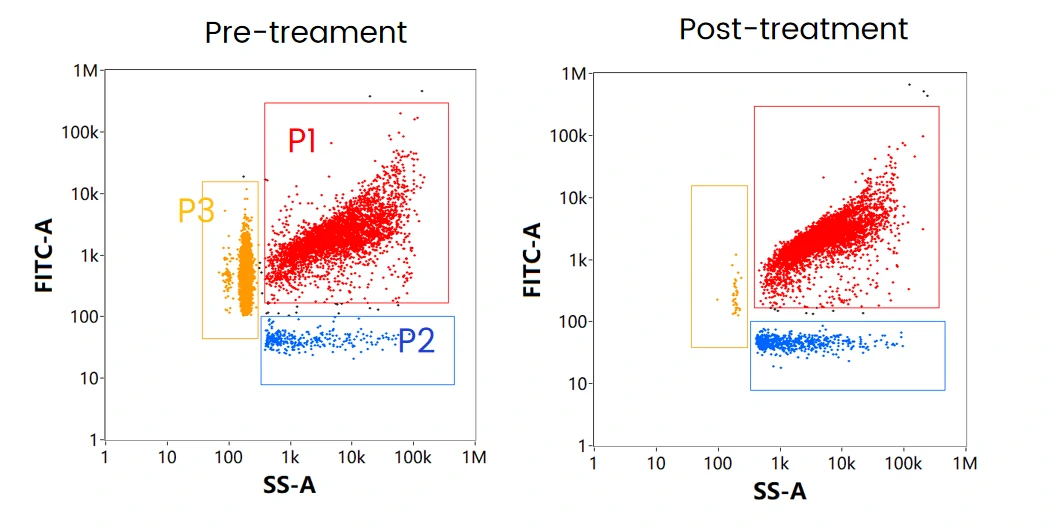
Nucleic Acid Localization Assay
The table summarizes the mRNA localization assay through different labeling strategies followed by nuclease treatment, which can be referred to optimize the formulating strategy and downstream process. By doing so, maximizing the effective loading of mRNA to obtain mRNA LNPs with high purity is possible.
Lipid Nanoparticles for Targeted siRNA Delivery
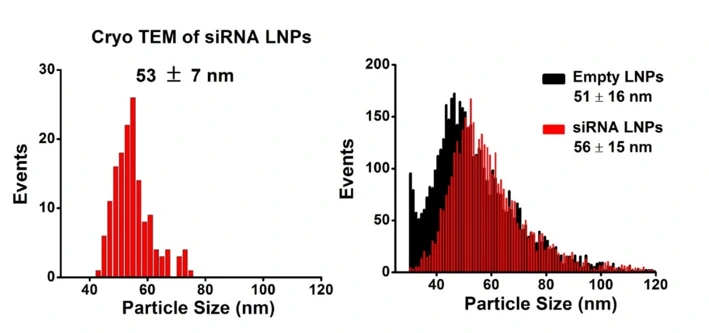
Complete discrimination between the empty and siRNA-loaded lipid nanoparticles is achieved based on SYTO 82 staining, the fraction of siRNA loading is determined to be approximately 100%. The size distribution of empty and siRNA-loaded LNPs are 51 and 56 nm through NanoFCM analysis, which is close to the cryo-EM measurement.
ACS Nano 2014, 8, 10998-11006
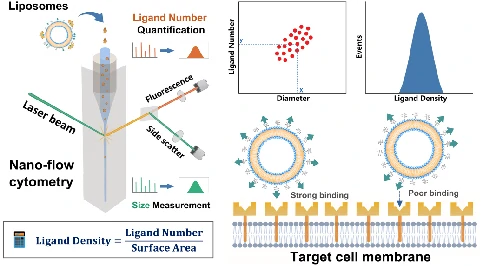
Functionalization with Targeting Ligands
The liposomes formulated by phospholipids, cholesterol, and a PEG layer were employed as model system. The development of a method for density quantification of targeting ligands at single-particle level was reported. This was achieved through the concurrent measurement of particle size and number of ligands on the same liposome by NanoFCM. By measuring the available ligand density of folate, it was identified that although the folate density kept increasing with the ligand input, the optimal density for cellular uptake fell in the range of 0.5 to 2.0 ligands per 100 nm2, which agreed well with the spike density of most viruses.
ACS Nano 2022, 16, 4, 6886–6897




