Clinical Diagnosis of Bacterial Infection and Resistance
It has been reported that individuals could be simultaneously infected with multiple strains of different susceptibility levels, and the population of resistant bacteria could be very low. However, if the minority population of resistant bacteria cannot be detected in time, an inappropriate prescription of antibiotics is usually the result. Therefore, detecting the minority population of antibiotic-resistant bacteria is crucial for clinical diagnosis. By employing a monoclonal antibody against TEM-1 β-lactamase and an Alexa Fluor 488-conjugated secondary antibody to selectively label resistant bacteria in green, and using the nucleic acid dye SYTO 62 to stain all the bacteria, the Flow NanoAnalyzer is able to detect and quantify as low as 0.1% of antibiotic-resistant bacteria. Furthermore, this approach is applied to detect antibiotic-resistant infection in clinical urine samples without cultivation, and the bacterial load of susceptible and resistant strains can be reliably quantified. This method provides a powerful tool for the fundamental studies of antibiotic resistance and holds the potential to offer rapid and precise guidance in clinical therapies.
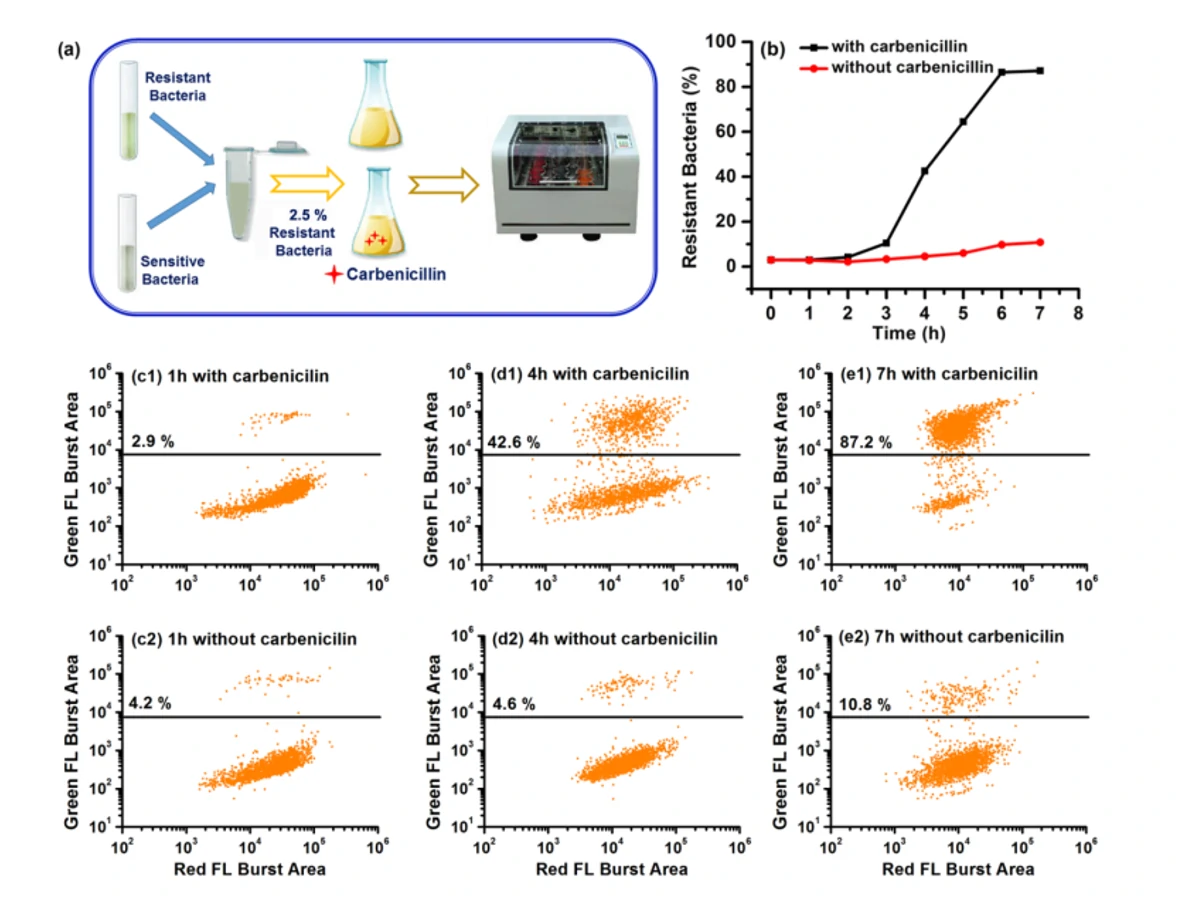
Figure 1. Tracking of the dynamic population change of antibiotic-resistant bacteria with and without antibiotics.
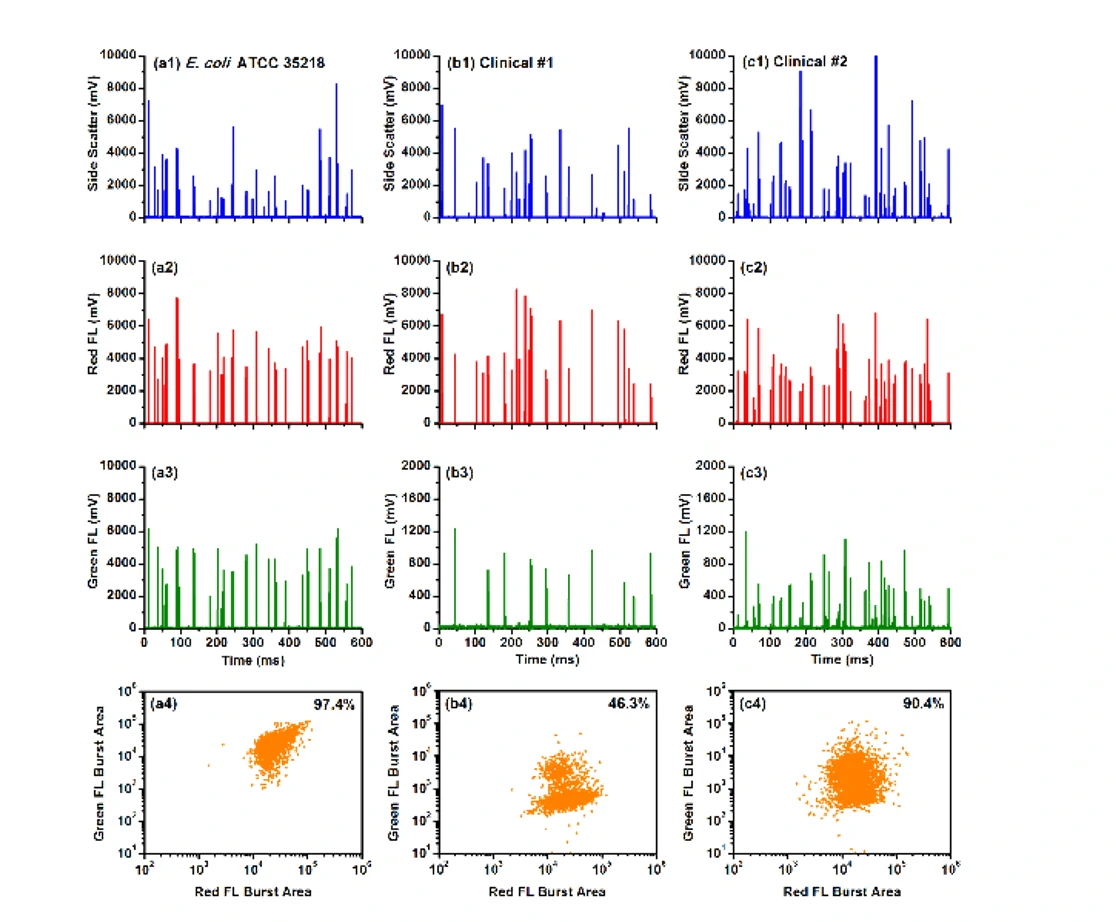
Figure 2. Analysis of E. coli ATCC 35218 (positive control) and two β-lactamase positive clinical urine samples upon dual fluorescent staining
Through fluorescent immunolabeling and nucleic acid staining of bacteria, detection of minority population of antibiotic-resistant bacteria is achieved.
Biosen. Bioelectron., 2016, 80, 323-330.




