Rapid Detection of Resistant Bacteria Based on β-lactamase Activity
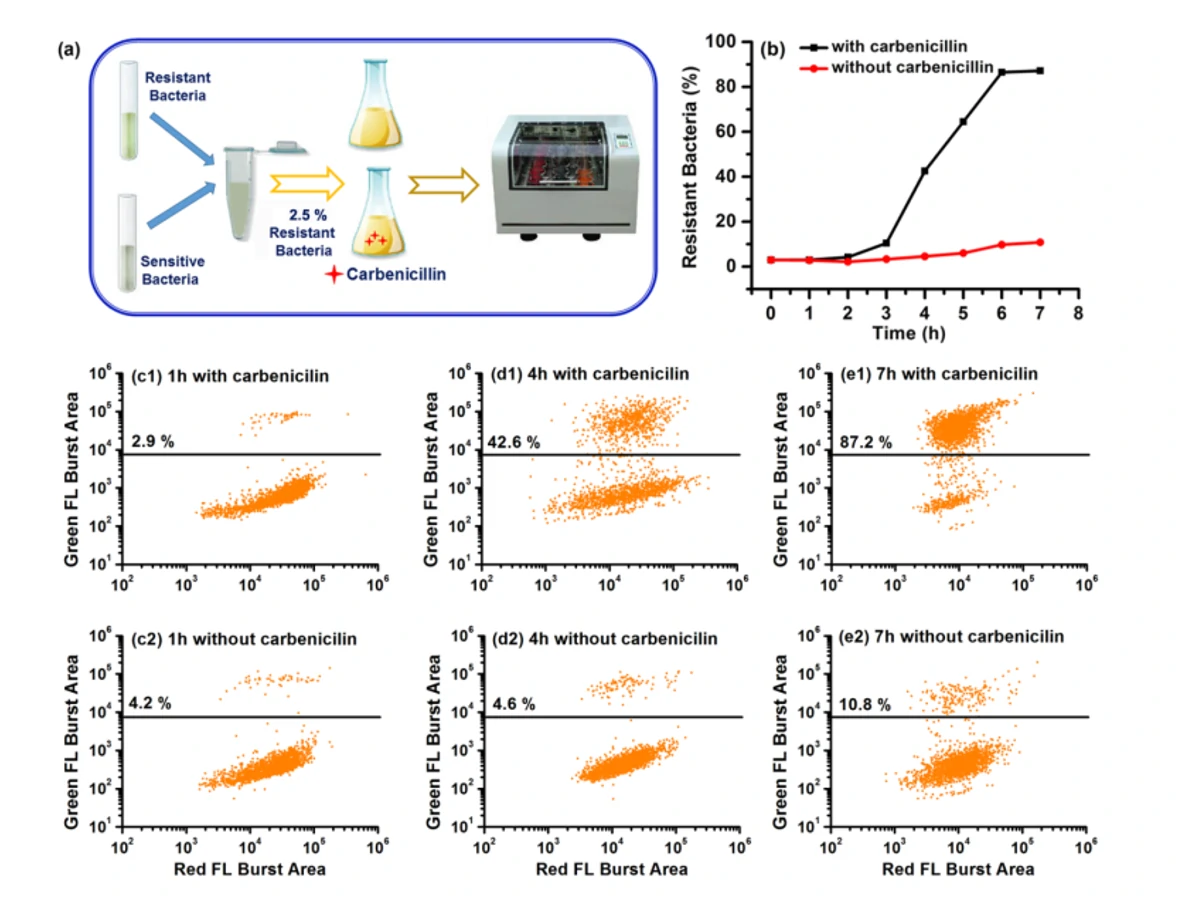
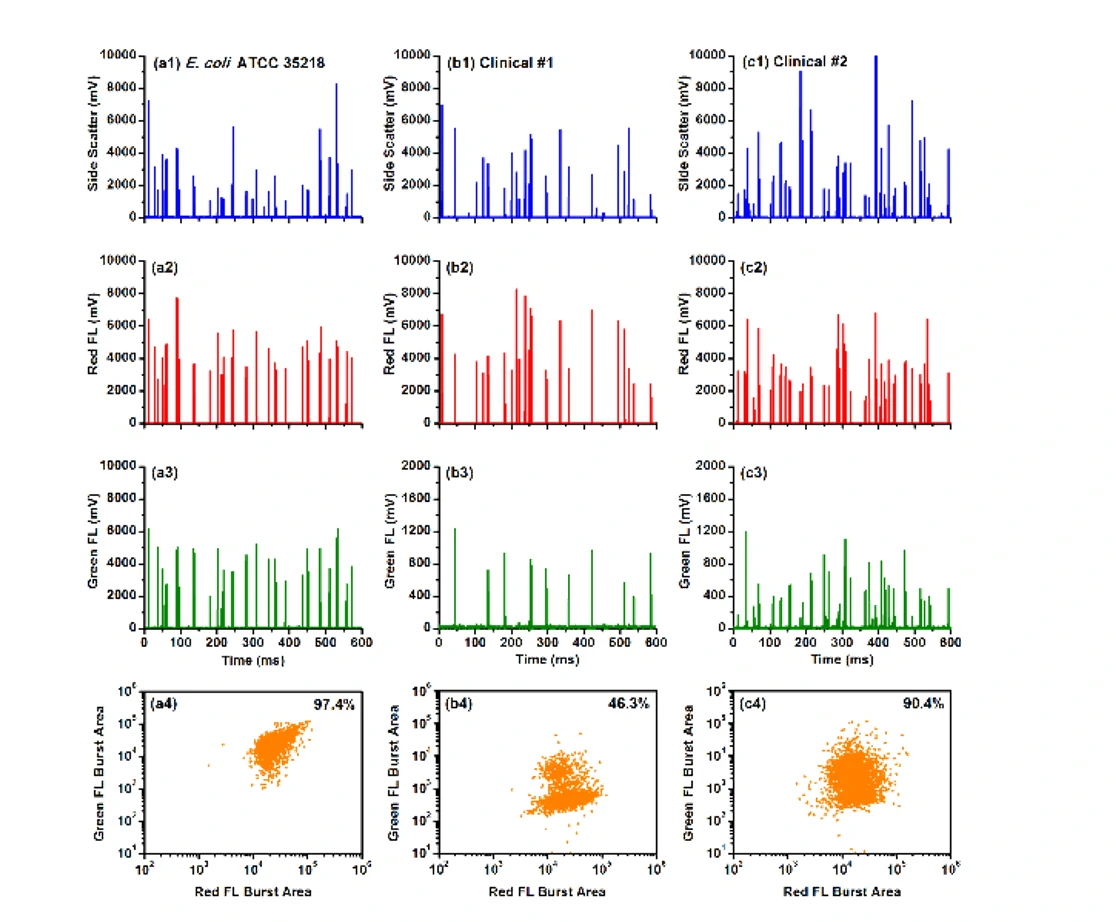
Bacterial resistance to antibiotics poses a significant clinical challenge in combating serious infectious diseases due to complicated resistance mechanisms and time-consuming test methods. Among the many molecular mechanisms that confer antibiotic resistance, the production of β-lactamase, which catalyzes the hydrolysis of β-lactam antibiotics, is a major and threatening mechanism. Additionally, it has been reported that individuals could be simultaneously infected with multiple strains of different susceptibility levels. Traditional detection methods cannot detect minority populations of antibiotic-resistant bacteria. Advanced tools are urgently needed to quickly diagnose antibiotic-resistant infections to initiate appropriate treatment. The hydrolyzed probes LBRL1 could attach to the enzyme, β-lactamase, and thus facilitate the covalent labeling of drug-resistant bacterial strains. Moreover, this β-lactamase-induced covalent labeling provides quantitative analysis of the resistant bacterial population (down to 5%) by the Flow NanoAnalyzer.
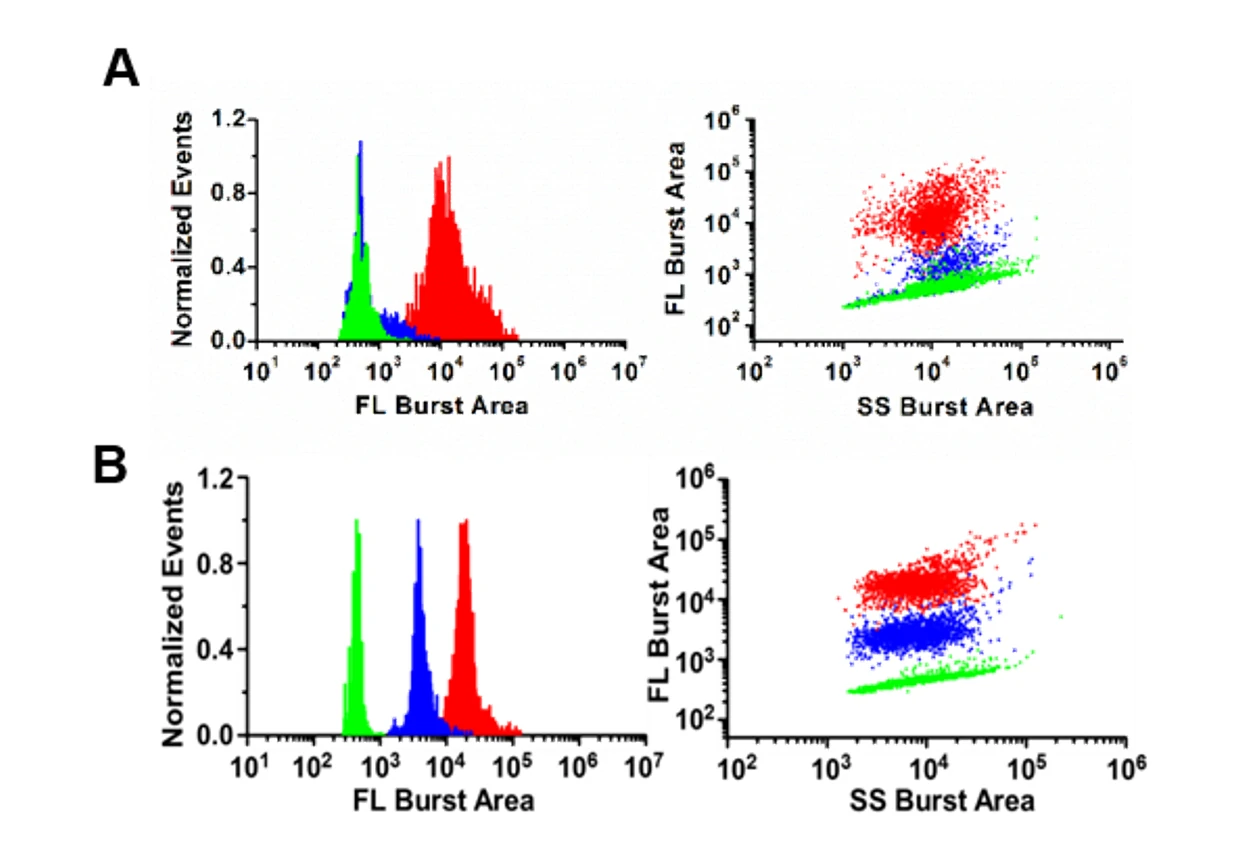
Figure 1. Analysis of bacteria resistance in single gram-negative bacterial cell.
(A) Bacteria are labeled with LBRL1.(B)Unlabeled B.
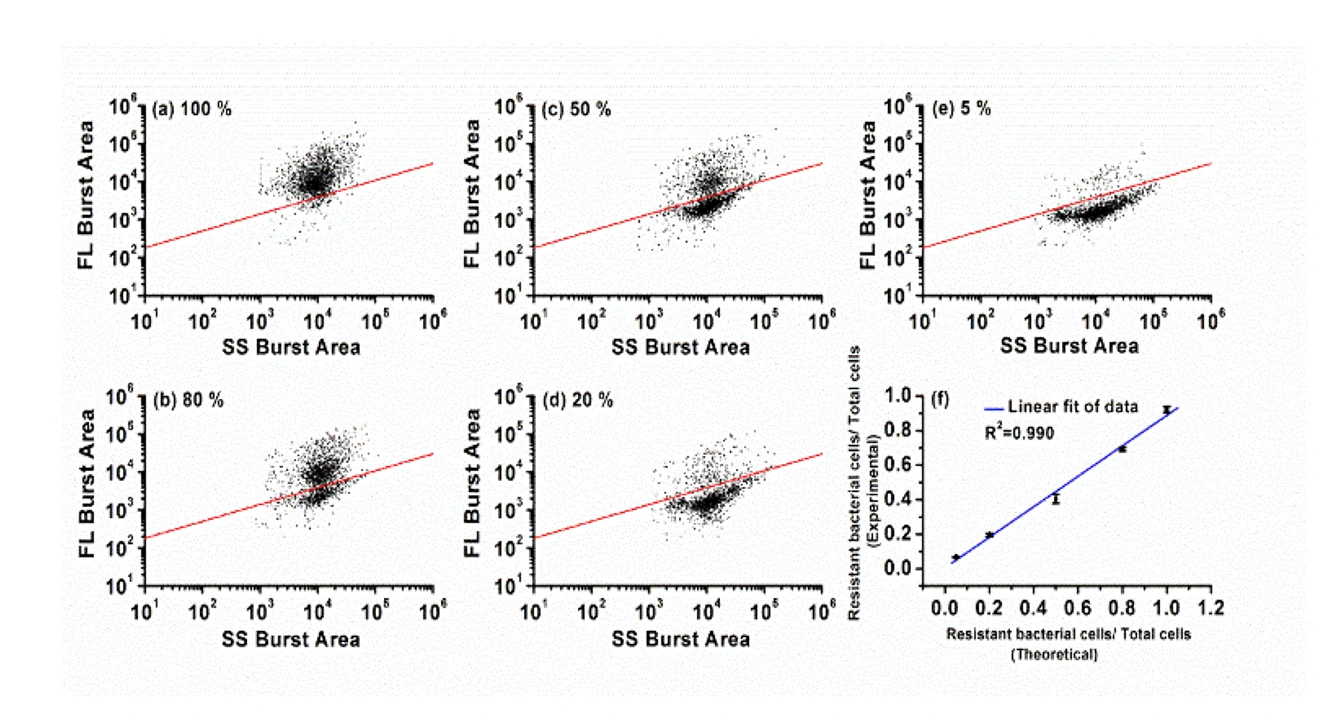
Figure 2. Differentiation of resistant E. coli JM109/pUC19 cells in bacterial mixtures.
The Flow NanoAnalyzer allows rapid single-cell detection and quantitative observation of the resistant bacterial population (down to 5%) through fluorescent probe LBRL1.
Chem. Eur. J., 2013, 19(33), 10903-10910.