Physical Properties: Size Distribution and Particle Concentration
EV content has been gaining increasing interest from the EV research community, primarily mRNAs, miRNAs and proteins. In addition, particular markers exposed on the lipid bilayers that determine specific interactions with target cells. However, it has been suggested that physical properties of the particles may also affect the behavior of EVs, such as the way they mediate intercellular-communication. In fact, in the case of engineered nanoparticles, it has been shown that their size may affect the uptake efficiency and kinetics, the internalization mechanism and also the subcellular distribution. Whether the size of natural vesicles might be also an essential factor that determines how easily they can diffuse in a tissue and how effectively cells can take them up is still unknown and represents the overarching question behind this study. Exosome size is reported to vary from 30-150 nm and, as a consequence of the small nature of the particles, an accurate size estimation tends to be elusive.

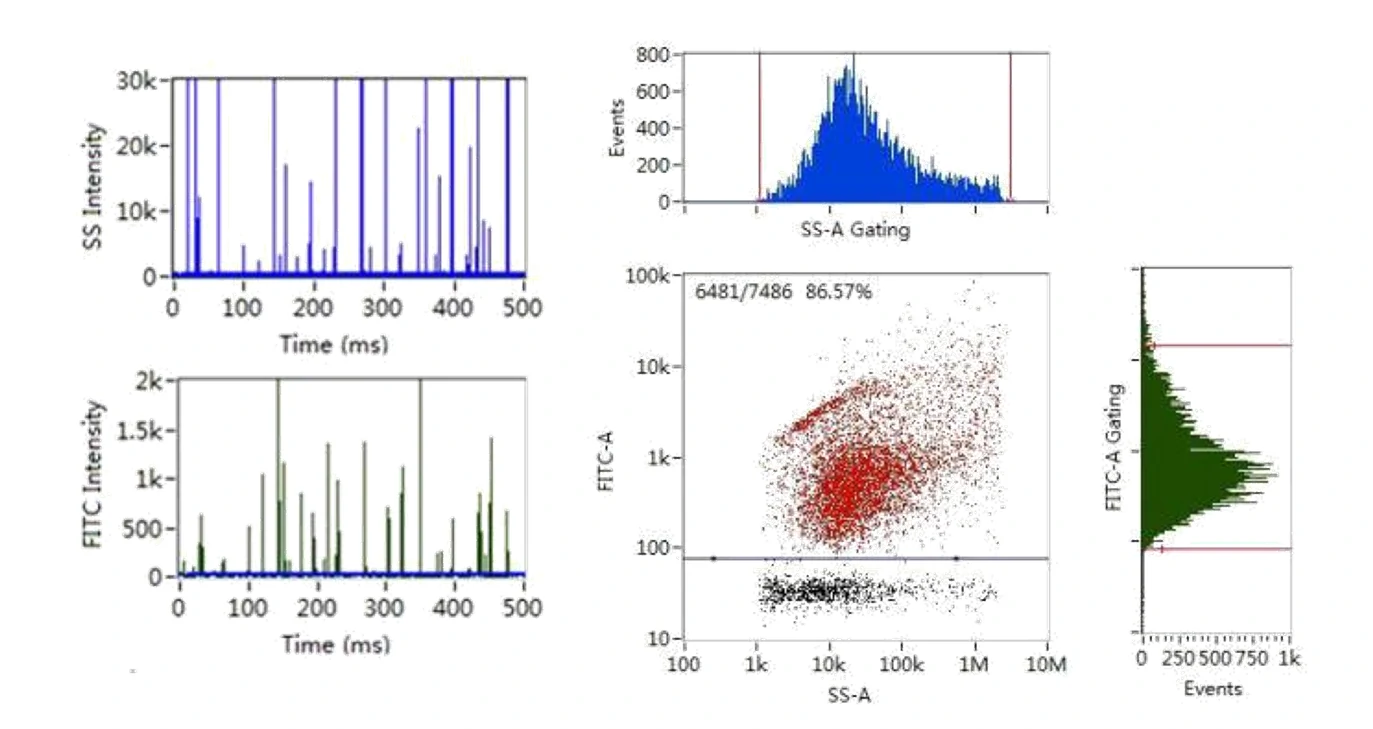
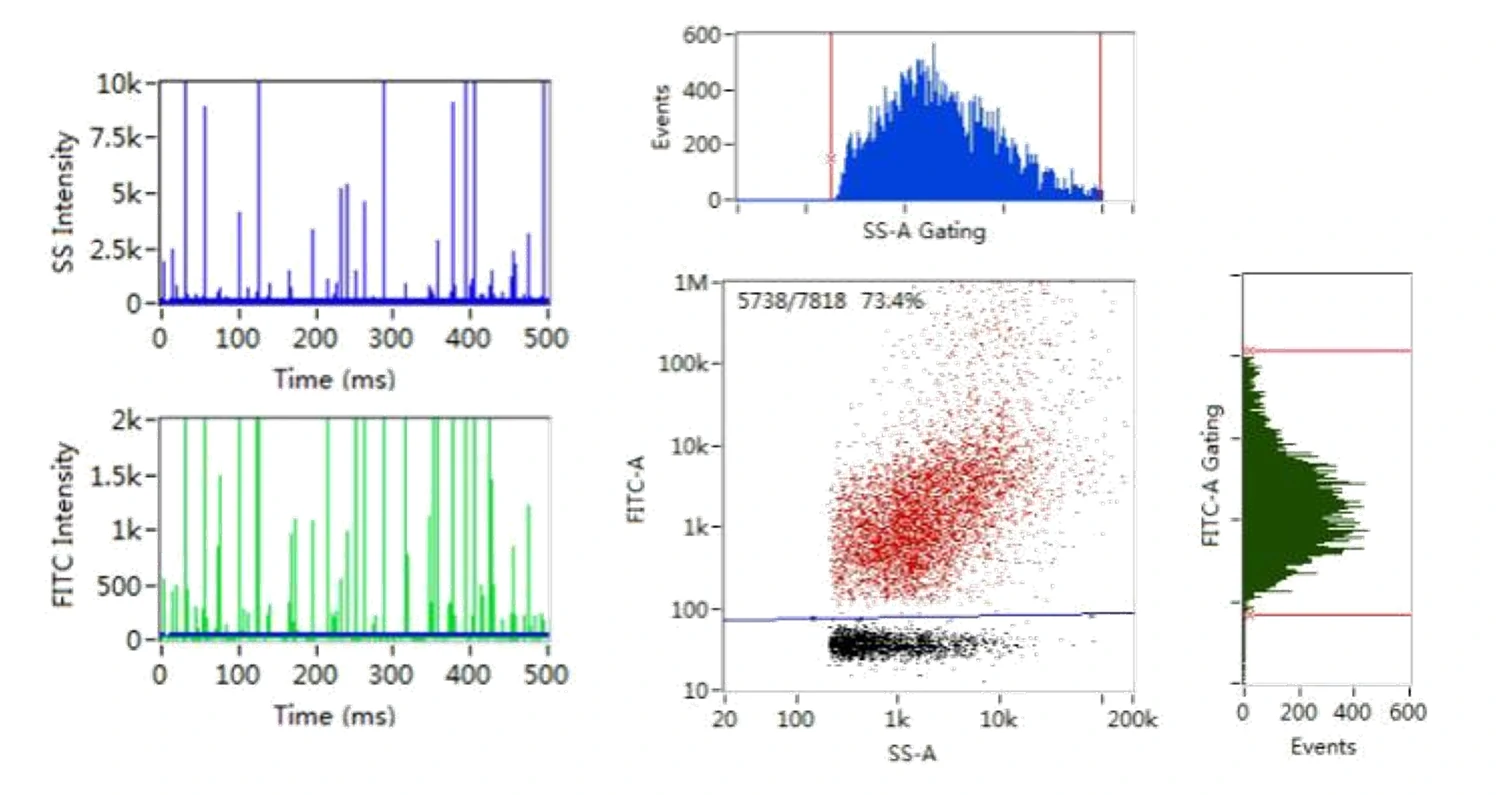
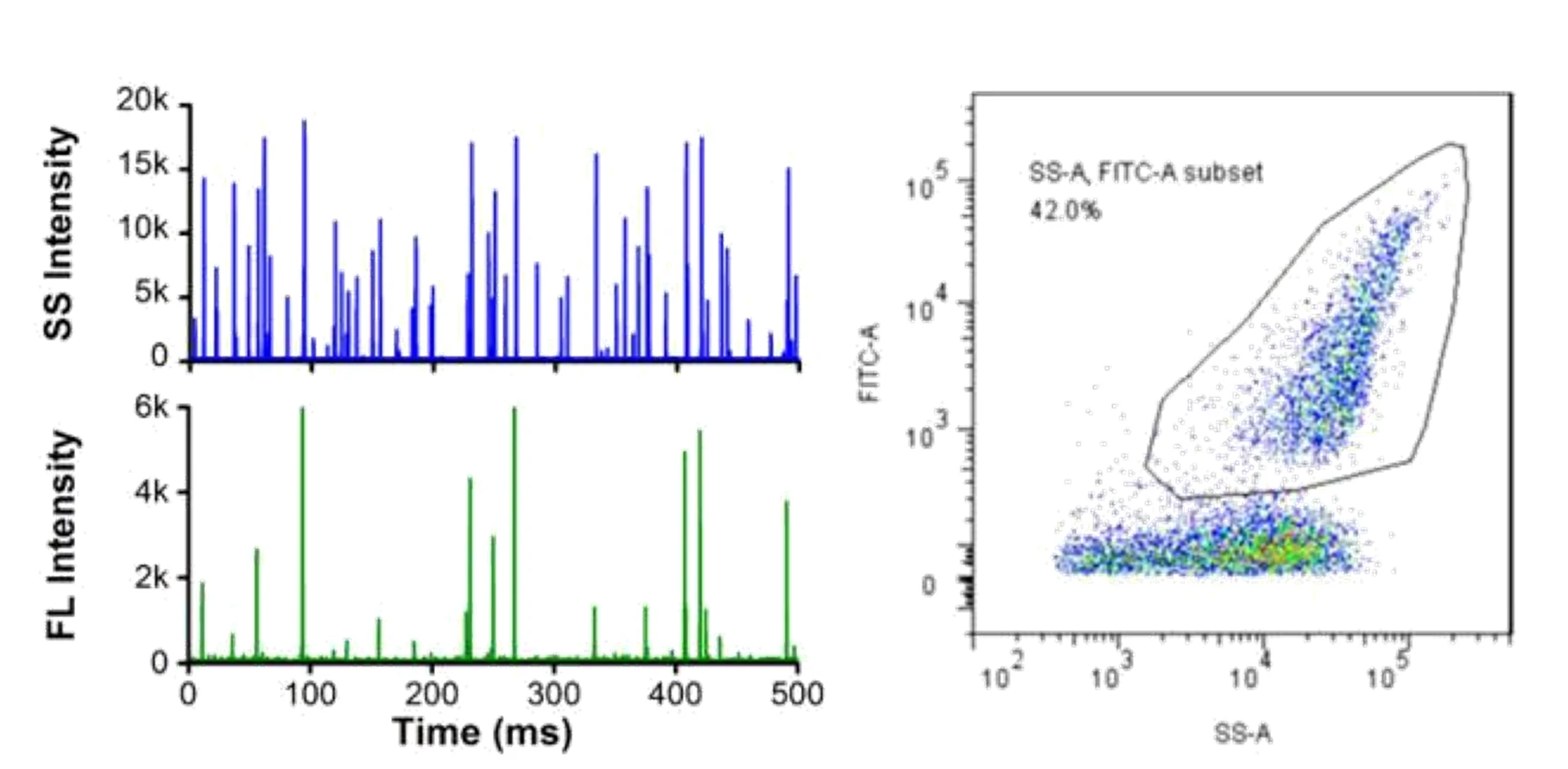
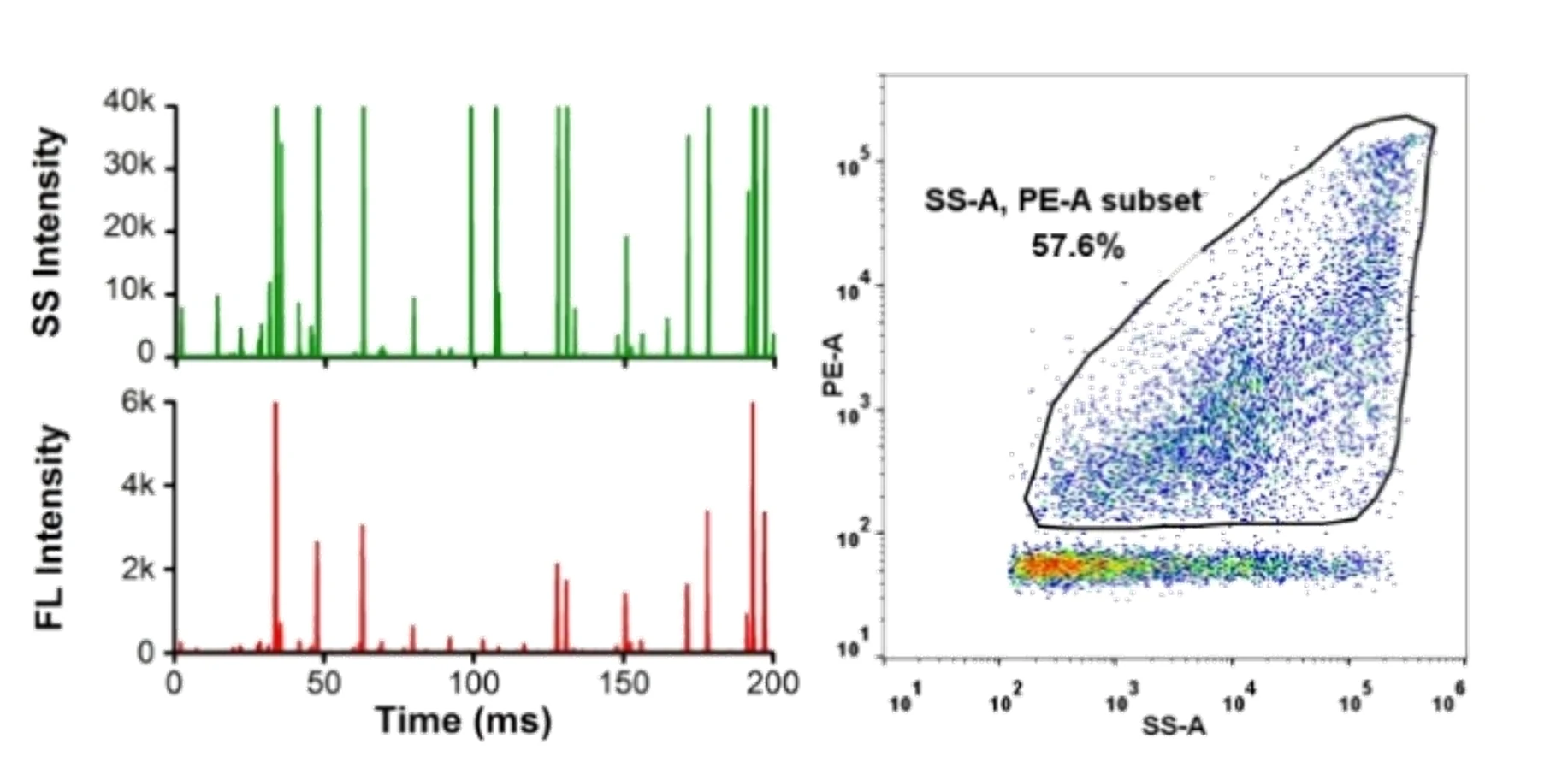
Here, monodisperse silica nanoparticles are used as the size reference standards. Therefore, the SS intensity of every single EV particle can be converted to particle size. Employing fluorescent nanoparticles of a known concentration as internal standard, the particle concentration of EVs can be acquired.
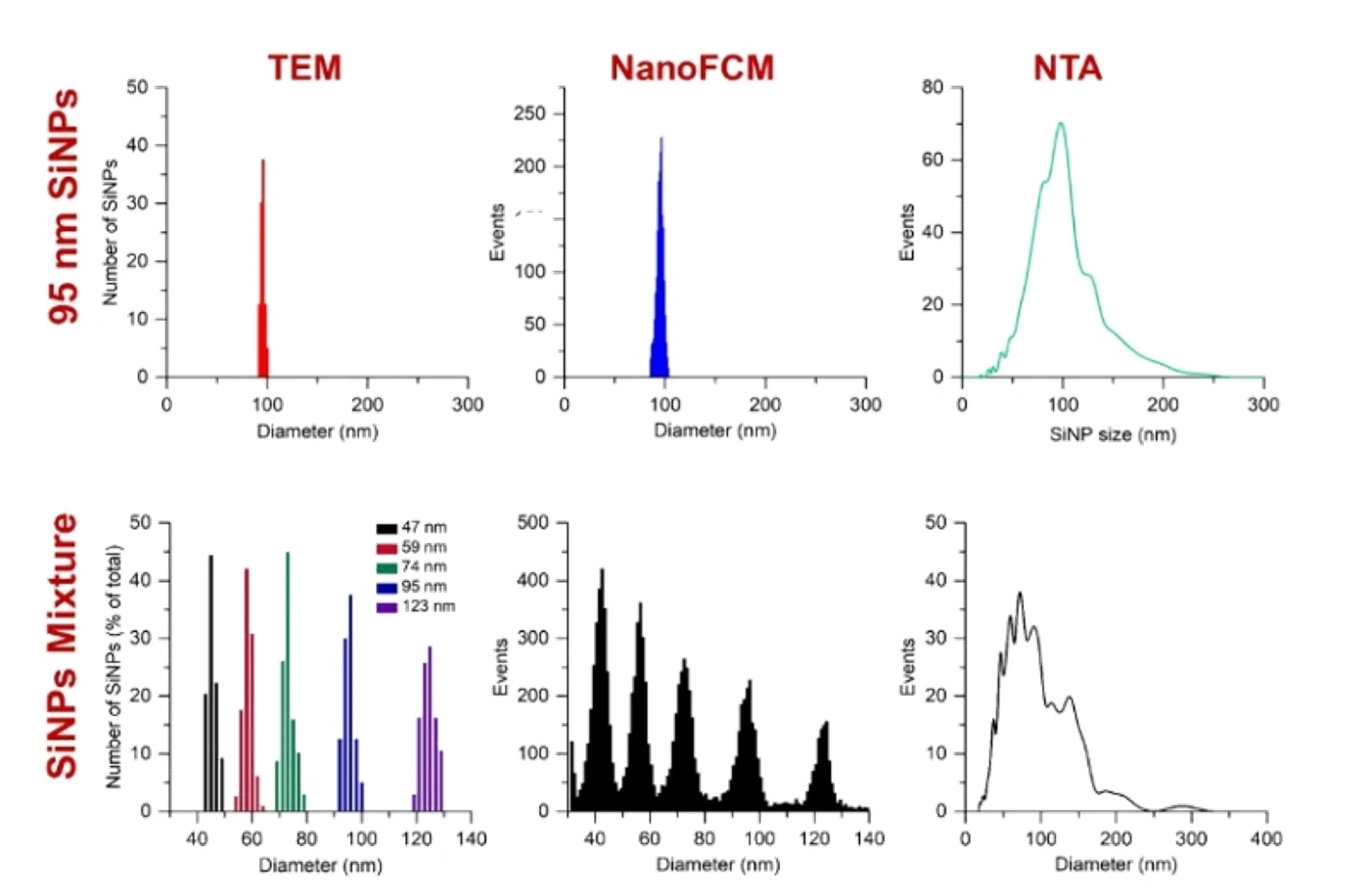
Figure 1. Comparison of TEM, Flow NanoAnalyzer, and NTA for particle size distribution analysis of silica nanoparticles.
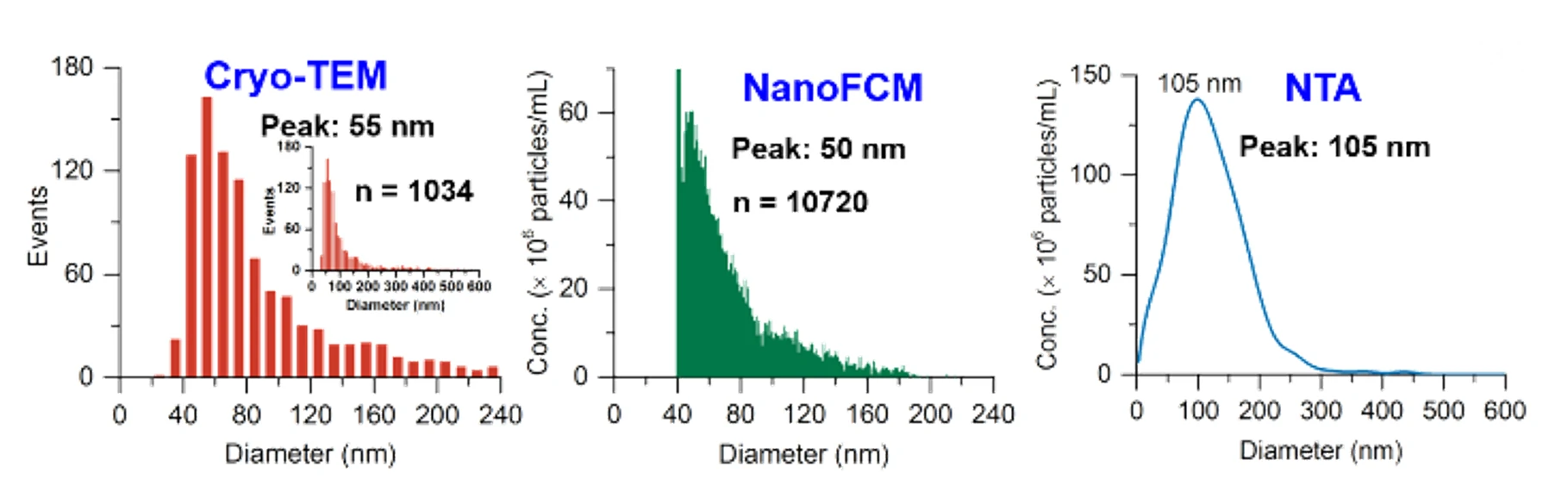
Figure 2. Size distribution analysis of EVs
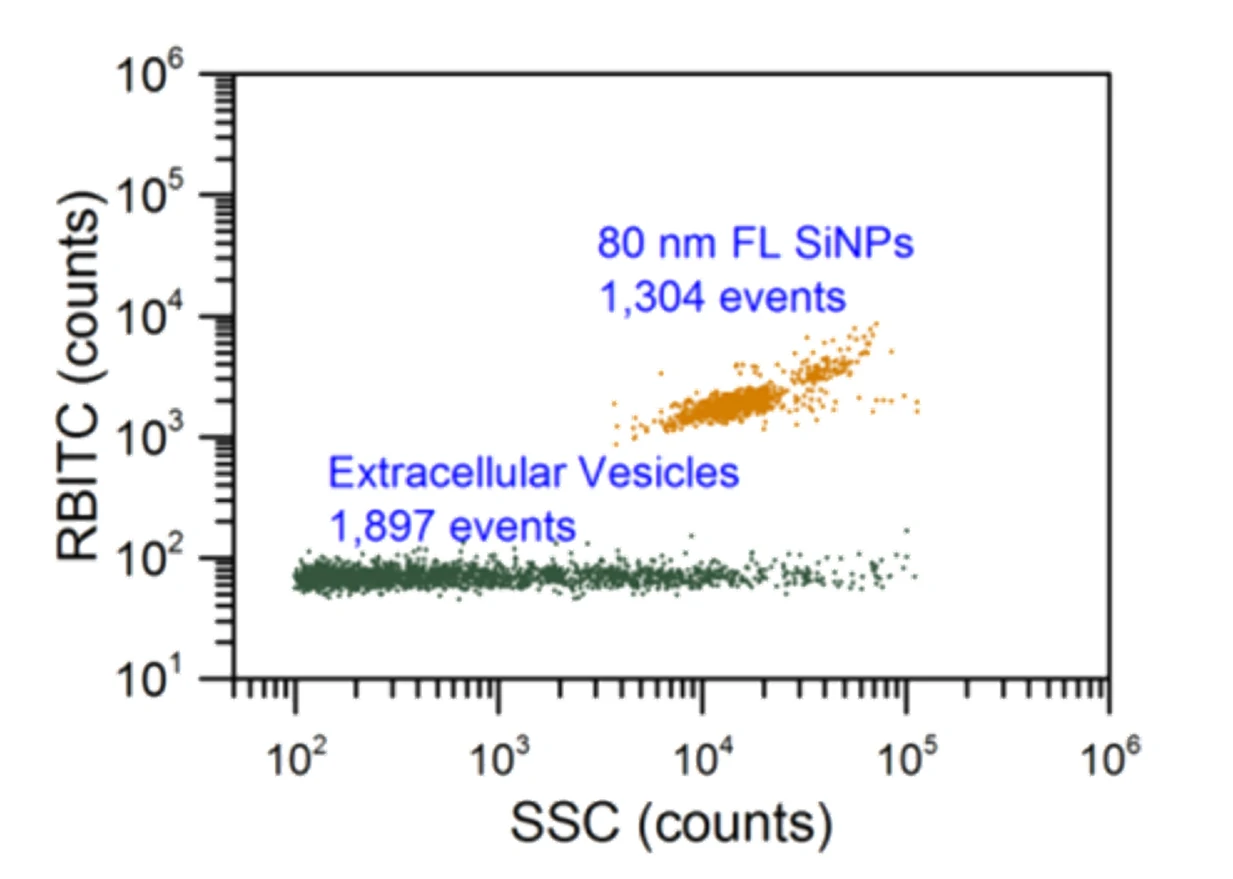
Figure 3. Particle concentration determination by Flow NanoAnalyzer.
ACS Nano, 2018, 12(1), 671-680.